We are committed to providing open-source software and tools for the scientific community. Our group Github organization can be found here. Below, we highlight a number of software tools from our publications. Older software tools that we believe have been fully supplanted by more recent ones are greyed out.
Research Themes: (select to filter)

Web App: https://askcos.mit.edu
Documentation: https://askcos-docs.mit.edu/
GitLab: https://gitlab.com/mlpds_mit/askcosv2
ASKCOS (Automated System for Knowledge-based Continuous Organic Synthesis) is an open source software suite hosting several data-driven chemistry tools for synthesis planning. It has seen use by 35,000+ chemists and is deployed at 15+ pharmaceutical and chemical companies. At the above link, we host a free-to-use web app deployment of ASKCOS.
predictive chemistry
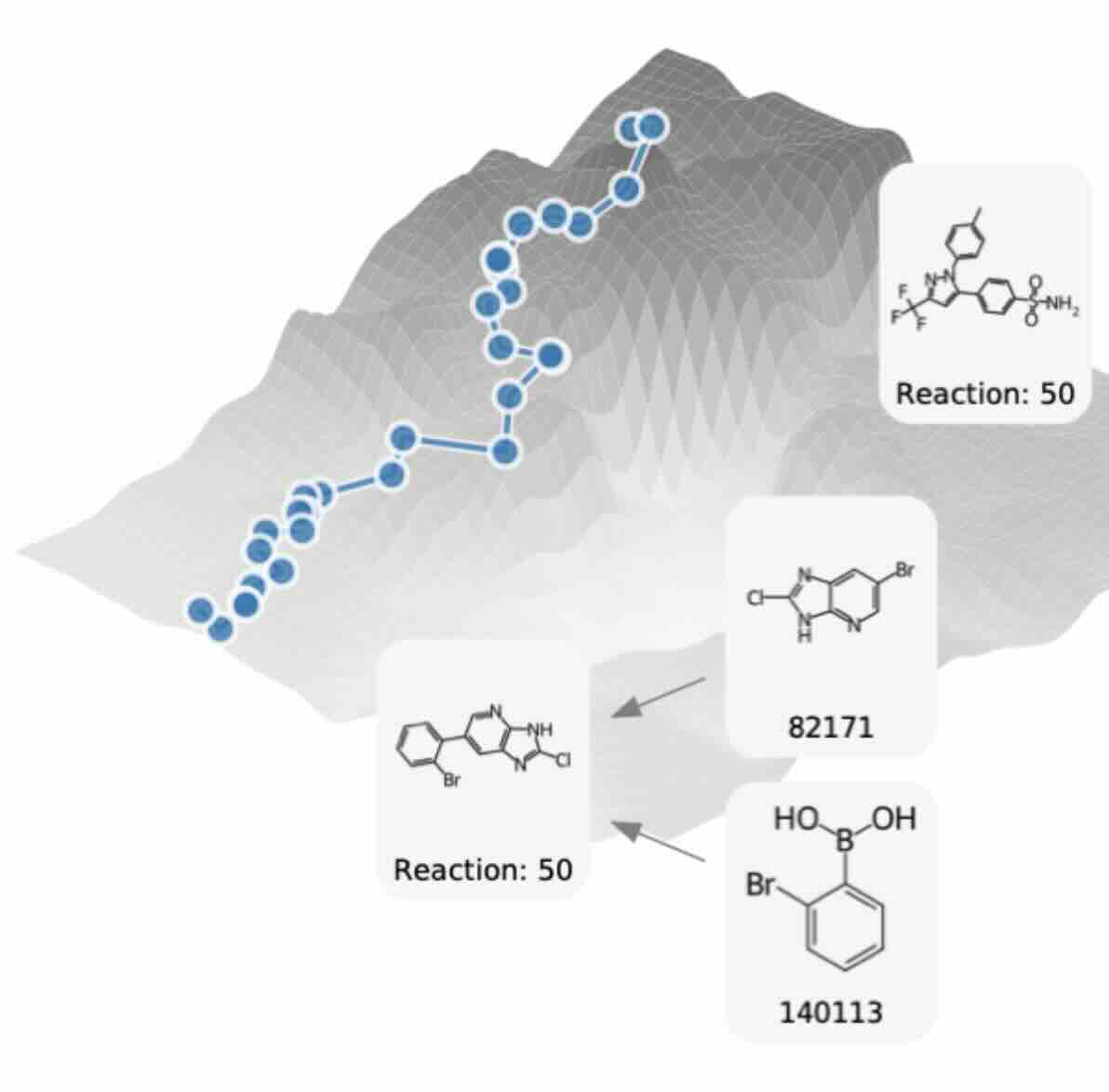
Website: https://prexsyn.readthedocs.io/en/latest/
GitHub: https://github.com/luost26/PrexSyn
PrexSyn is an efficient, accurate, and programmable framework for synthesizable molecular design that simultaneously generates molecules and their synthesis routes. PrexSyn excels at “projecting” molecules into the synthesizable chemical space and sampling molecules that optimize a scoring function while satisfying property queries. PrexSyn was trained on post-fix notations paired with molecular properties at billion-scale using only two GPUs and 32 CPU cores in two days with PrexSyn Engine, a real-time, high-throughput C++-based data generation pipeline.
design and optimization
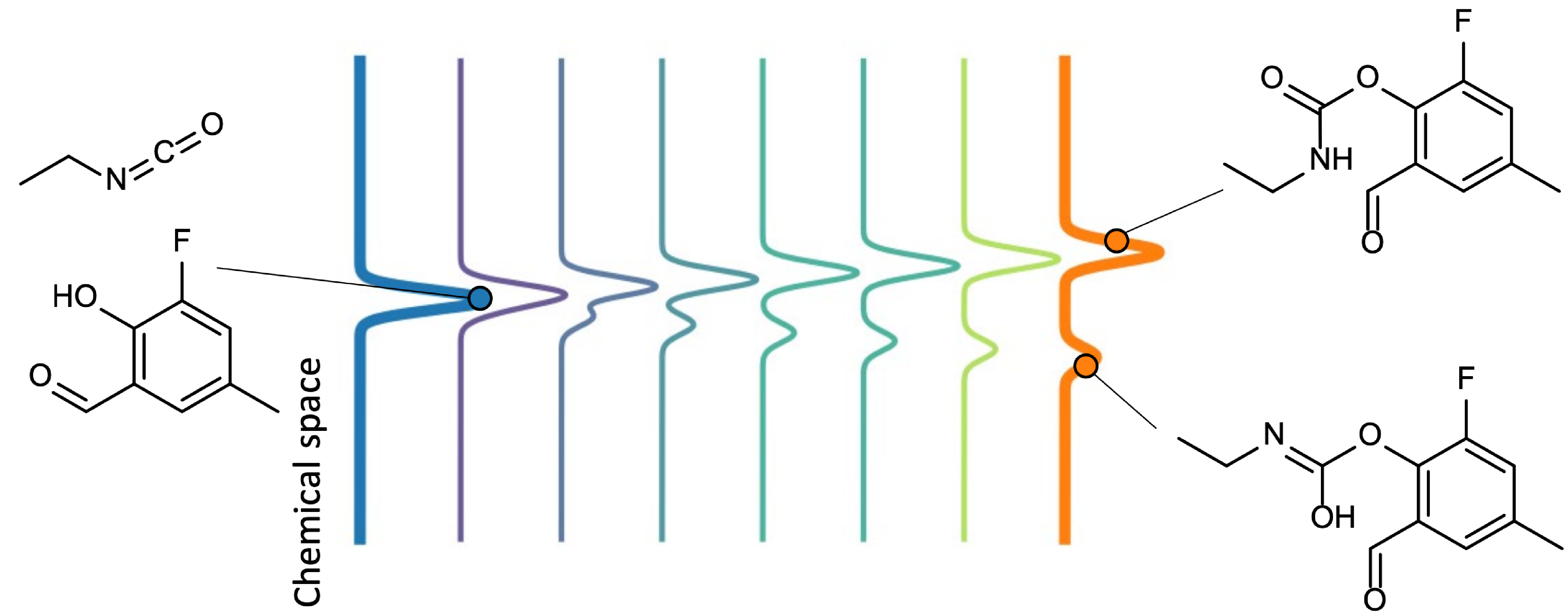
GitHub: https://github.com/FongMunHong/FlowER
FlowER uses generative flow matching to model chemical reaction as a process of electron redistribution, conceptually aligns with arrow-pushing formalisms. It aims to capture the probabilistic nature of reactions with mass conservation where multiple outcomes are reached through branching mechanistic networks evolving in time.
molecular representationpredictive chemistry

GitHub: https://github.com/coleygroup/ms-pred
ICEBERG is a geometric deep learning model that simulates collision-induced dissociation in mass spectrometry to generate chemically plausible fragments and their relative intensities with awareness of collision energies and polarities. We utilize ICEBERG predictions to facilitate structure elucidation by ranking a set of candidate structures based on the similarity between their predicted in silico MS/MS spectra and an experimental MS/MS spectrum of interest.
metabolomics

GitHub: https://github.com/coleygroup/diffms
DiffMS is a generative discrete graph diffusion model for structure elucidation from mass spectra. It is a formula-restricted encoder-decoder generative network that achieves state-of-the-art performance on this task. The encoder utilizes a transformer architecture and models mass spectra domain knowledge such as peak formulae and neutral losses, and the decoder is a discrete graph diffusion model restricted by the heavy-atom composition of a known chemical formula.
metabolomicsmolecular representation
GitHub: https://github.com/coleygroup/ms-pred
MARASON is a retrieval-augmented mass spectrum simulator with neural graph matching. We retrieve reference structures and spectra from a reference library based on similarity to the target structure. It is a novel model that incorporates neural graph matching to enhance a fragmentation-based neural network to improve Retrieval Augmented Generation.
metabolomics
GitHub: https://github.com/samgoldman97/mist-cf
MIST-CF (Metabolite Inference with Spectrum Transformers - Chemical Formula) is an extension of the MIST framework that infers precursor chemical formulae directly from tandem mass spectra in a de novo setting. Using a transformer-based energy model, it ranks candidate formulae without requiring reference spectral databases.
metabolomics
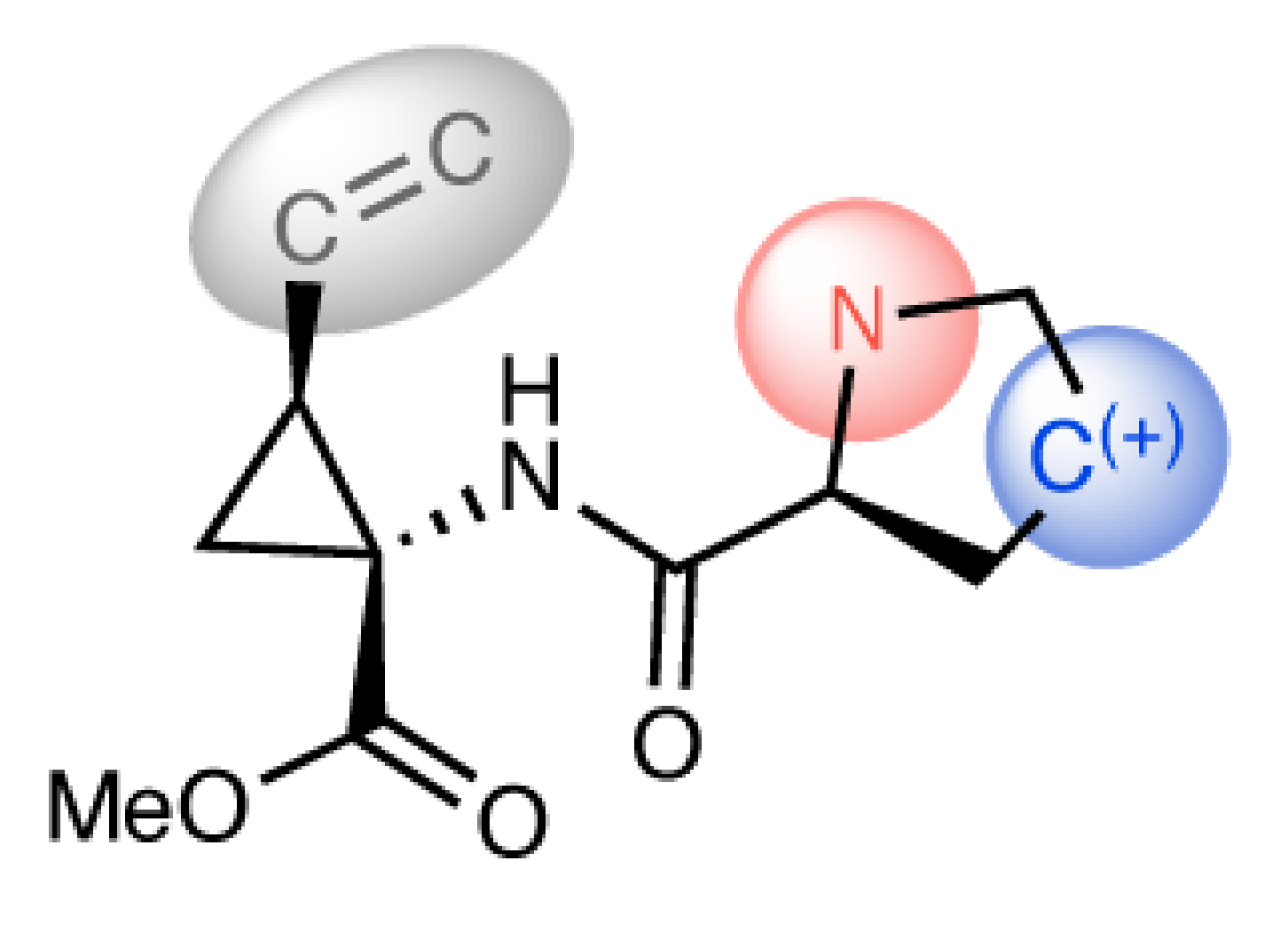
GitHub: https://github.com/jihye-roh/higherlev_retro
Higher-Level Retrosynthesis introduces a novel framework for computer-aided retrosynthesis that focuses on the broader retrosynthetic strategy. We abstract the detailed substructures in the intermediates of a synthetic pathway that do not appear in the target product, allowing our algorithm to emphasize the general, higher-level strategies instead of considering the specifics of chemically-equivalent functional groups.
predictive chemistry
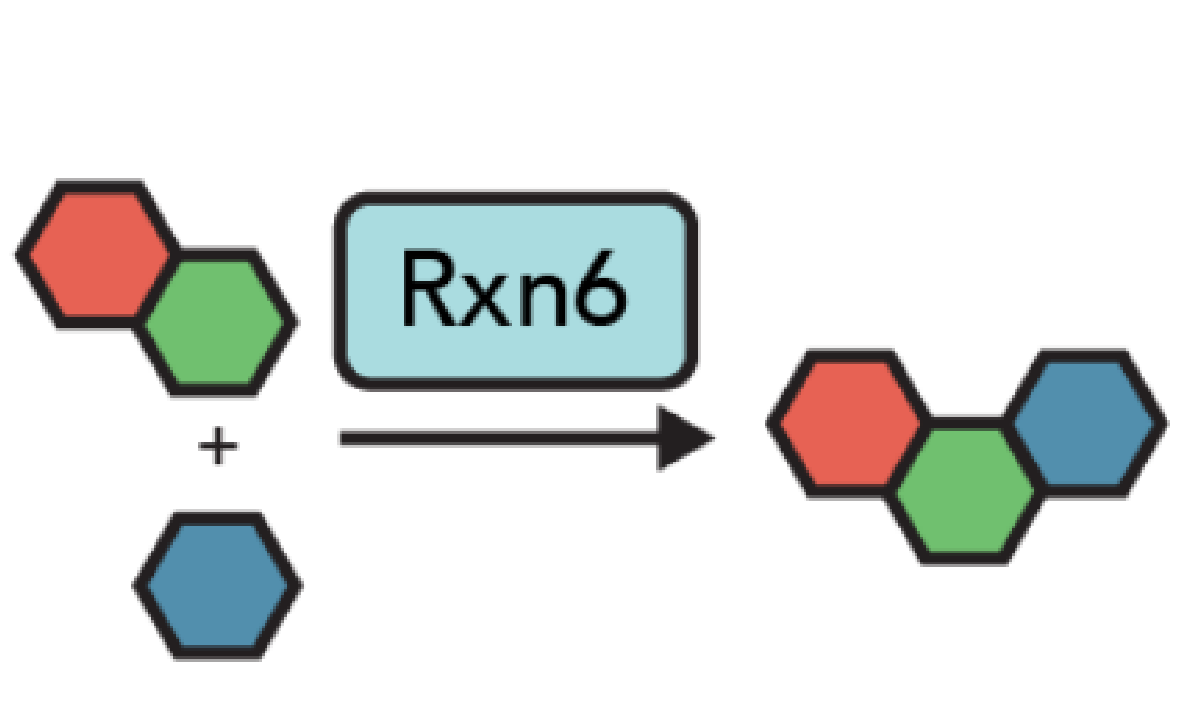
GitHub: https://github.com/coleygroup/synformer
SynFormer is a generative artificial intelligence model for navigating synthesizable chemical space. It is designed to generate molecules along with their synthetic routes in a scalable manner. It enables (1) local chemical space exploration, where the model generates synthesizable analogs of a reference molecule, and (2) global chemical space exploration, where the model aims to identify optimal molecules according to a black-box property prediction oracle.
design and optimizationmolecular representation

GitHub: https://github.com/coleygroup/shepherd
ShEPhERD is a generative diffusion model for generating molecules conditioned on explicit representations of shape, electrostatics, and pharmacophores. It is applicable to bioisosteric drug design such as natural product ligand hopping, bioactive hit diversification, and bioisosteric fragment merging.
molecular representationdesign and optimization
GitHub: https://github.com/connorcoley/rdchiral
RDChiral is a tool for RDKit’s RunReactants to improve stereochemistry handling.
molecular representationpredictive chemistry

GitHub: https://github.com/coleygroup/rdcanon
RDCanon is a package designed for canonicalizing SMARTS and Reaction SMARTS templates. It reorders SMARTS to optimize querying speed such as for database search. This optimization is invariant of atom mapping.
datapredictive chemistry

GitHub: https://github.com/coleygroup/virtual_flask
The Virtual Flask is a cheminformatic application designed to generate reaction networks given a set of initial molecules and a corpus of templates and enables reaction prediction. A suite of supporting tools facilitate the analysis and visualization of generated chemical reaction data. This guide provides detailed instructions for setting up and running the Virtual Flask locally for development purposes. Both the Corpus and Analysis dashboards can be hosted locally and run simultaneously.
automationpredictive chemistry
GitHub: https://github.com/coleygroup/sparrow
A workflow to simultaneously select molecules and their synthetic routes for lead optimization and design-make-test cycles. This optimization approach aims to minimize synthetic cost while selecting the molecules that are most likely to fulfill design constraints. More details about SPARROW can be found in our preprint.
design and optimizationpredictive chemistry
GitHub: https://github.com/jenna-fromer/qPO
qPO (multipoint Probability of Optimality) is an acquisition strategy for discrete optimization that is motivated by pure exploitation. It maximizes the probability that the batch includes the true optimum, which is expressed as the sum over individual acquisition scores and thereby circumvents the combinatorial challenge of optimizing a batch acquisition function
predictive chemistrydesign and optimizationautomation
GitHub: https://github.com/itai-levin/easie
EASIE (Exploration of chemical Analog Space, Implicitly and Explicitly) is a computational workflow to (1) score and select retrosynthetic pathways on the basis of perceived route diversifiability, (2) estimate property distributions of the resulting enumerated space to inform or constrain reactant selection, and (3) enumerate synthetic pathway-constrained analogs via selection of suitable alternative reactants.
predictive chemistry
GitHub: https://github.com/wenhao-gao/mol_opt
PMO (Practical Molecular Optimization) is a open-source benchmark to facilitate the transparent and reproducible evaluation of algorithmic advances in molecular optimization. This repository supports 25 molecular design algorithms on 23 tasks with a particular focus on sample efficiency (oracle calls).
design and optimization
GitHub: https://github.com/coleygroup/pyscreener
A pythonic interface to high-throughput virtual screening software. It was designed to have a minimal interface under the principal that a high-throughput virtual screen is intended to be a broad strokes technique to gauge ligand favorability. It supports Vina, Quick-Vina, Smina, Psovina, and DOCK6.
automationmolecular representation
GitHub: https://github.com/coleygroup/molpal
MolPAL is a software for the accelerated discovery of compounds in high-throughput virtual screening environments. Bayesian optimization algorithms for docking-based virtual screening decreases the computational cost of using docking to identify the majority of top-scoring compounds in virtual libraries by over an order of magnitude, aided by surrogate machine learning models that prioritize the screening of molecules that are associated with better docking scores.
design and optimization
GitHub: https://github.com/coleygroup/Graph2SMILES
Graph2Smiles is a graph-to-sequence model that predicts the SMILES string of a reaction product given a reaction template and a reactant molecule for one-step retrosynthesis.
predictive chemistry
GitHub (ROGI): https://github.com/coleygroup/rogi
GitHub (ROGI-XD): https://github.com/coleygroup/rogi-xd
ROGI is a quantitative metric for describing the roughness of molecular property landscapes, inspired by fractal dimension, which helps characterize the presence of activity cliffs and correlates with the out-of-sample error of machine learning models in molecular property prediction. ROGI-XD extends this framework to evaluate the roughness of learned representations from chemical foundation models, enabling assessment of how well these models capture the geometric complexity of molecular property landscapes.
molecular representation
GitHub: https://github.com/coleygroup/polymer-chemprop
Polymer Chemprop adapts the Chemprop message-passing neural network architecture for predicting polymer properties. It supports training and evaluation workflows tailored to polymer datasets, enabling accurate property prediction and materials design.
molecular representationpredictive chemistry
GitHub: https://github.com/qai222/libsyn_tools
libsyn_tools is a set of Python utilities for designing and analyzing library synthesis campaigns, including enumeration, plate mapping, and data processing workflows.
automationdesign and optimization
GitHub: https://github.com/wuRoy/CVRP_pipette_scheduling
CVRP Pipette Scheduling formulates high-throughput liquid-handling operations as a capacitated vehicle routing problem to minimize execution time on multi-channel pipetting robots.
automation
GitHub: https://github.com/coleygroup/QM-augmented_GNN
QM-augmented GNN integrates quantum-mechanical descriptors with graph neural networks to improve molecular property prediction and reaction-barrier estimation. The repository provides code to featurize molecules with QM calculations and train augmented models.
molecular representationpredictive chemistry
GitHub: https://github.com/samgoldman97/ms-pred
SCARF (Prefix-Tree Decoding for Predicting Mass Spectra from Molecules) uses an intermediate strategy for predicting mass spectra from molecules by treating mass spectra as sets of molecular formulae. After first encoding an input molecular graph, we decode a set of molecular subformulae, each of which specify a predicted peak in the mass spectrum, the intensities of which are predicted by a second model. The model has been depreciated in favor of a new model “ICEBERG”.
metabolomics
GitHub: https://github.com/wenhao-gao/SynNet
SynNet is an amortized approach to generate synthetic pathways as a Markov decision process conditioned on a target molecular embedding. This approach allows us to conduct synthesis planning in a bottom-up manner and design synthesizable molecules by decoding from optimized conditional codes. The approach leverages neural networks to probabilistically model the synthetic trees, one reaction step at a time, according to reactivity rules encoded in a discrete action space of reaction templates. The model has been depreciated in favor of a new model “SynFormer”.
design and optimizationmolecular representation
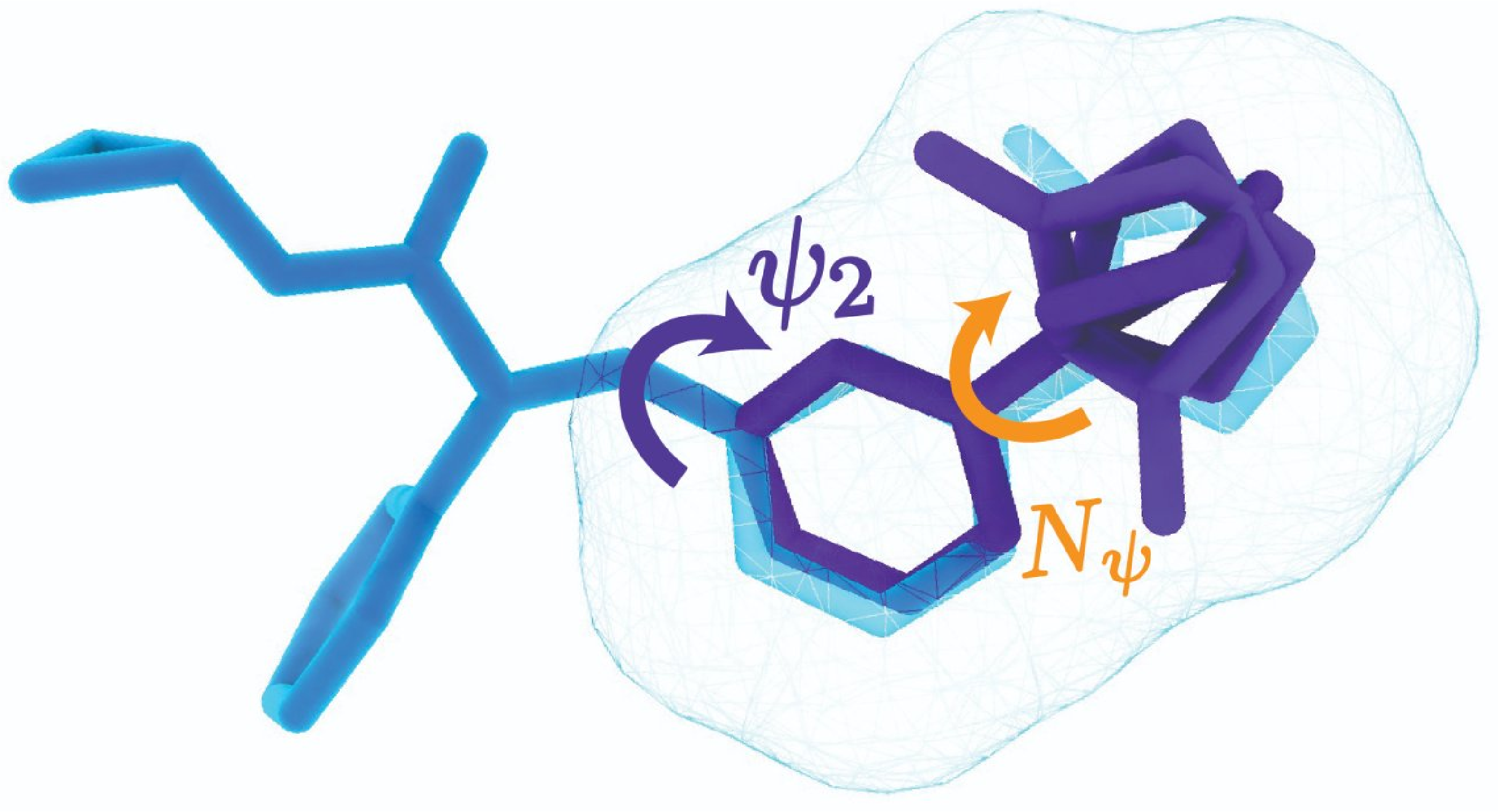
GitHub: https://github.com/keiradams/SQUID
SQUID is a multimodal 3D generative model that enables shape-conditioned 3D molecular design by equivariantly encoding molecular shape and variationally encoding chemical identity. We ensure local geometric and chemical validity of generated molecules by using autoregressive fragment-based generation with heuristic bonding geometries, allowing the model to prioritize the scoring of rotatable bonds to best align the growing conformational structure to the target shape. The model has been depreciated in favor of a new model “ShEPhERD”.
molecular representationdesign and optimization
Website: https://open-reaction-database.org
GitHub: https://github.com/Open-Reaction-Database
The Open Reaction Database (ORD) is a community-driven repository and schema for storing and sharing machine-readable organic reaction data to accelerate synthesis planning, reaction prediction, and experiment design.
datapredictive chemistry
GitHub: https://github.com/chemprop/chemprop
ChemProp v2 is a message-passing neural network framework for molecular property prediction that supports modern architectures, uncertainty estimation, and interpretability for applications in drug discovery and materials science.
molecular representationpredictive chemistry

Website: https://tdcommons.ai
GitHub: https://github.com/mims-harvard/TDC
TDC is an open-science platform providing machine-learning-ready datasets, benchmarks, and leaderboards covering diverse therapeutic modalities and stages of drug discovery, facilitating reproducible and scalable AI research.
datapredictive chemistrydesign and optimization
GitHub: https://github.com/CrystalEye42/OpenChemIE
OpenChemIE is an open-source toolkit for chemistry information extraction, providing models and pipelines to automatically identify and convert chemical structures, reactions, and entities from text and figures in scientific literature.
datamolecular representation